Nuclear Magnetic Resonance Spectroscopy (NMR)
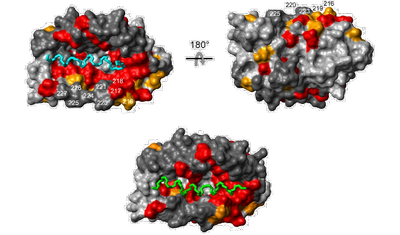
- The α-helices of the KIS domain (green) and the Kv4.3 N terminus (cyan) can bind to the same surface pocket on the KChIP core structure as shown by mapping of NMR chemical shift perturbation data onto a surface representation of KChIP.
Practically, NMR studies of proteins imply
- the expression and purification (mg amounts) of suitably isotope-labeled (15N, 13C, 2H) proteins for heteronuclear NMR experiments and
- the recording and processing of NMR spectra (1D, 2D, 3D, …) followed by
- a sequence-specific resonance assignment (protein backbone and side chains) or other types of spectral analysis.
Our institute is equipped with a Bruker Avance 600 NMR spectrometer with a cryoprobe allowing the investigation of proteins up to a size of 30-40 kDa at relatively low sample concentrations (< 300 µM).
3D structure determination by NMR requires NOE assignment and the measurement of additional structural constraints such as J coupling constants, residual dipolar couplings and paramagnetic relaxation enhancement (PRE). These data are then used as input for structure calculations which result in a family of structures.
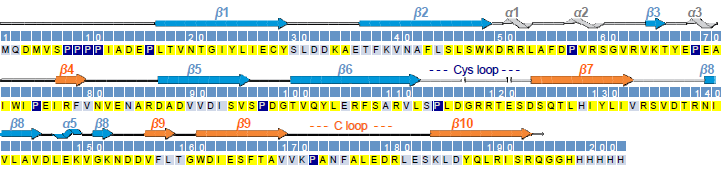
- Sequence-specific assignment and secondary structure of the extracellular domain of GLIC (yellow: assigned, light blue: unassigned, dark blue: proline).
Recent examples of our work are
- the analysis of a potassium channel-interacting protein (KChIP) that revealed the structural basis for control of surface expression of Kv4 channel complexes by an autoinhibitory domain in the KChIP4a subunit [Schwenk et al., J Biol Chem., 2008].
- the determination of the structural and dynamical properties of the monomeric extracellular domain of a prokaryotic nAChR homologue from the bacterium Gloeobacter violaceus [Chasapis et al., Biochemistry, 2011].


 Previous:
Mass Spectrometry
Previous:
Mass Spectrometry
